Error when importing all terminologies
|
Hello Jiba,
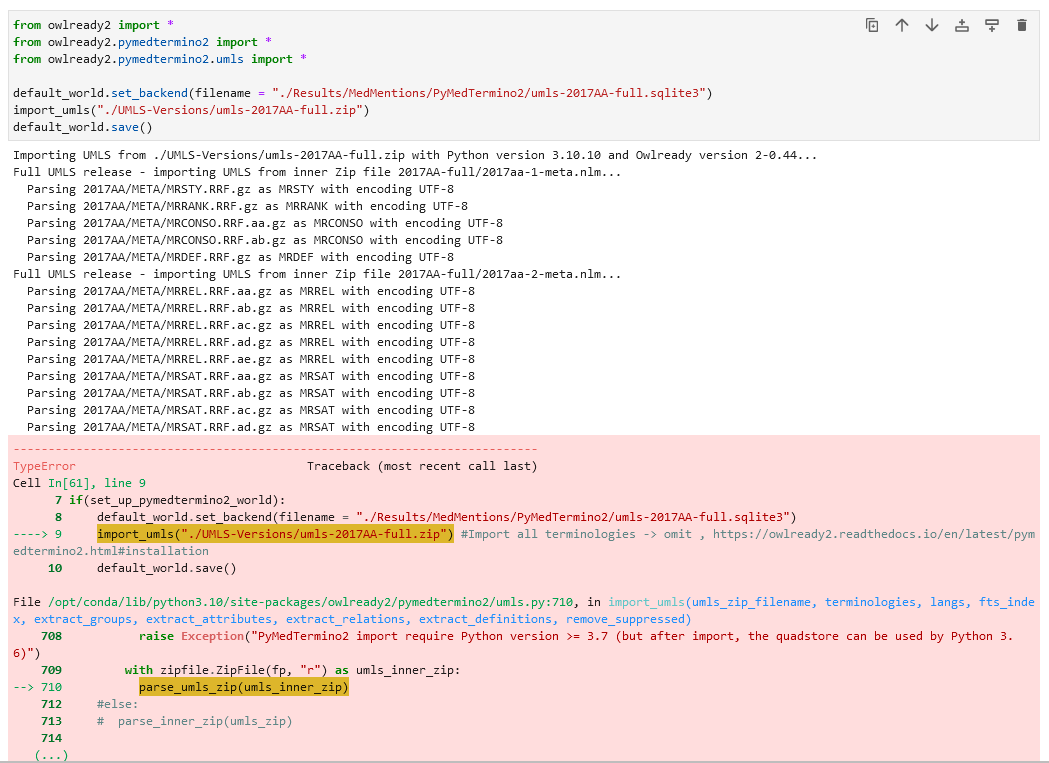
I would like to make use of the MedMentions dataset by gathering all synonyms for each CUI in the dataset. MedMentions was created 2017 so I am using the umls-2017AA-full release. Based on the PyMedTermino2 installation instructions, I set up PyMedTermino2 with the terminologies "ICD10", "SNOMEDCT_US", "CUI" as shown in the documentation (https://owlready2.readthedocs.io/en/latest/pymedtermino2.html#installation). But unfortunately errors occured, when I tried to query several CUIs. For instance the CUI "C0847557" with CUI['C0847557']can not be found and returns: CUI["C0847557"] # None I checked for a handful of concepts, for which the querying failed, in which terminologies they are: Terminologies with some concepts for which querying failed: - ICPC2P (C0847557) - NCI (C1708520) - CPM (C1254354) - CHV · AOD (C0683579, C0680954) - ICD10AM (C0845989) - CHV · MDR · MDRSPA · MDRDUT · MDRFRE · MDRGER · MDRITA · MDRJPN (C0858354) - MEDCIN (C3646020) My assumption is that only CUIs of concepts occuring in any of the defined terminologies are imported (e.g. if only SNOMED-CT is provided as a terminology, then only CUIs linking to concepts in SNOMED-CT are stored and retrievable? (Was not clear to me based on the documentation but makes sense) So that would mean I need to import all terminologies, since I don't know beforehand to which terminology a CUI belongs to. I went ahead and omitted the terminologies parameter according to the instructions to load all terminologies, but unfortunately, after a while of letting "import_umls" run (created file is 31.9 GB large) and processing the UMLS zip, it fails with following error: 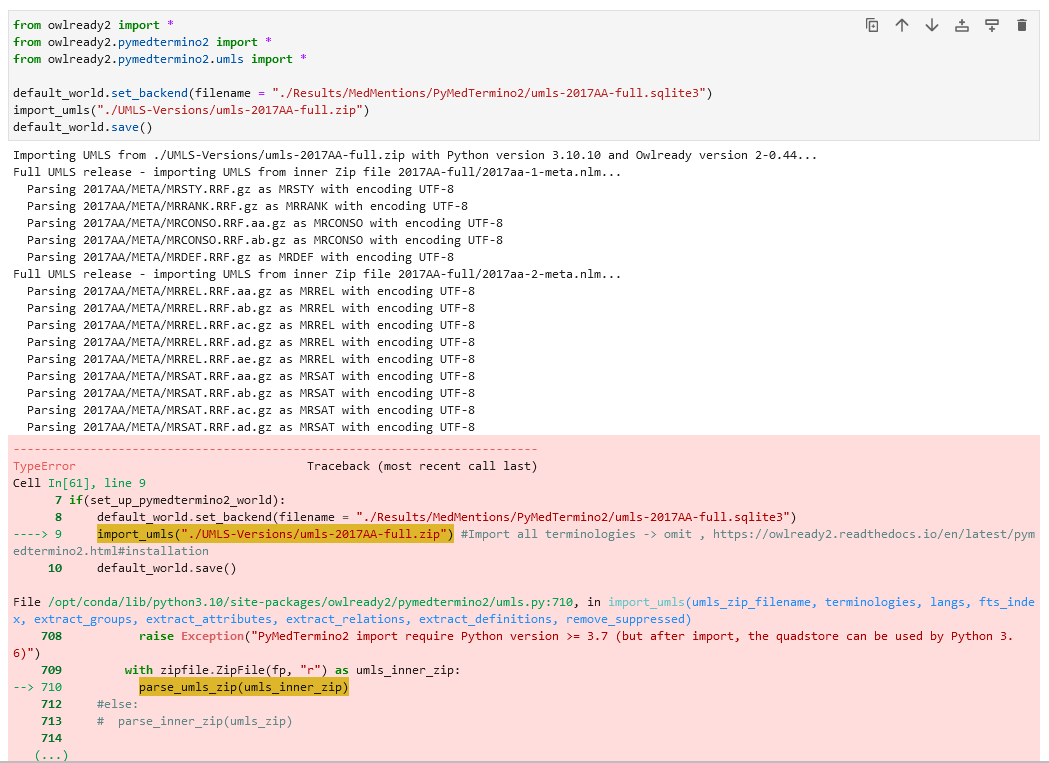 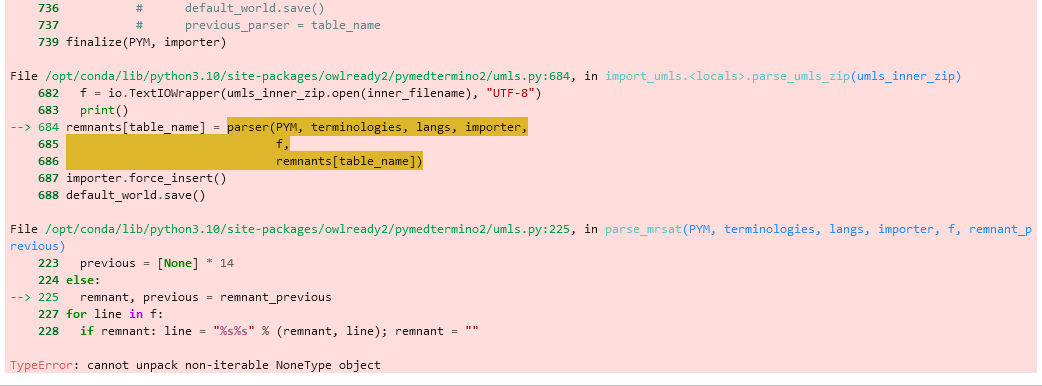 How can I fix this issue and load PyMedtermino2 properly with all terminologies? Kind regards Kim Tang |
|
Okay, so gathered synonyms for a lot of CUIs and was wondering why fewer error messages (exceptions) occured as compared to when I gathered the labels for each CUI.
Turns out the CUIs are apparently stored in PyMedTermino2 even if the respective terminology is not imported, but the information seems incomplete. All the concepts from above don't have a label, but they might have accessable synonyms that can be queried: 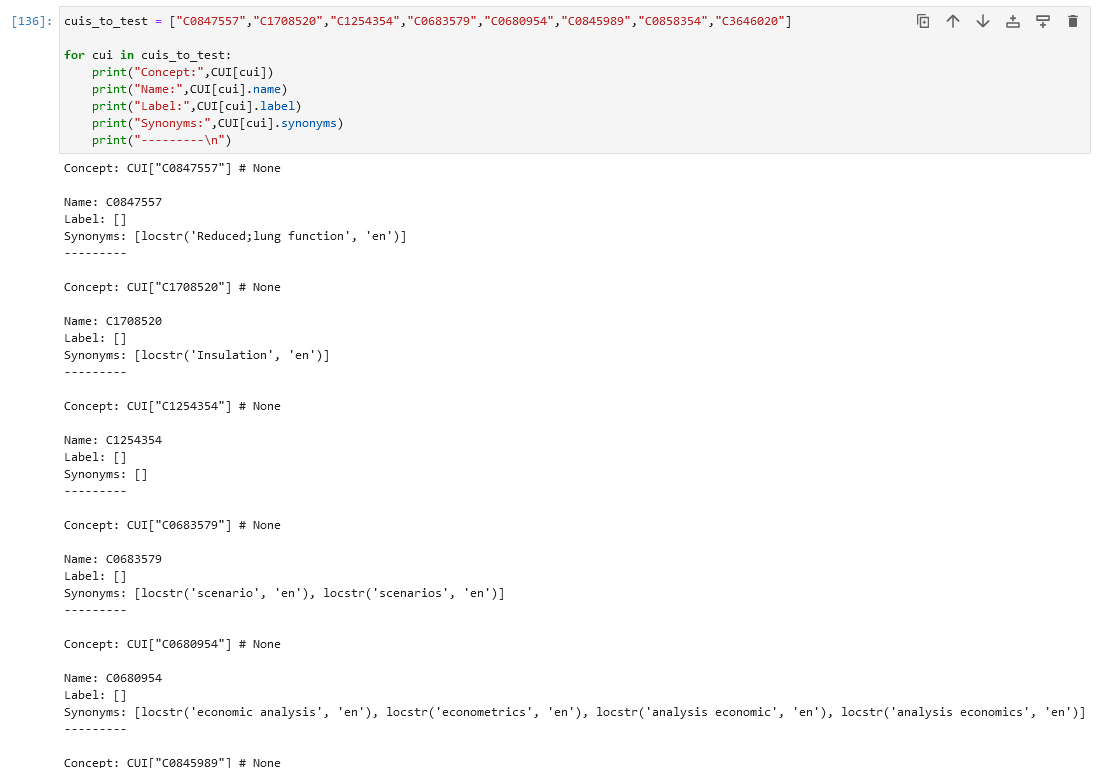 These synonyms don't stem from any imported terminology though (ICD10, SNOMED, CUI). I still dont understand why this is the case, but perhaps it helps when looking into it, if it turns out to be some error. Any help is appreciated! :) |
|
Administrator
|
Actually, the CUI imported are those from the imported terminology + those from the UMLS semantic types. The CUIs without label that you observed where extracted from MRSTY and correspond to semantic types.
If you want to know from where a CUI was extracted, you can use the .originals attribute that returns the list of terms in the original terminology. If it is empty, the CUI comes from semantic types only. Jiba |
|
Hello Jiba,
thank you for the clarification! And do you know why the initial error occured as outlined above, when I tried to import all terminologies (by omitting the terminology parameter)?
|
|
Hello -
Was this (umls) terminology import issue ever resolved? I am trying to import UMLS 2024AA full zip file, running in to a similar error, wondering if anyone has worked around this issue? Thank you, much appreciated! ---------------------On Python 3.10.12, Ubuntu 22 (WSL2)---------------------------------------- Full UMLS release - importing UMLS from inner Zip file 2024AA-full/2024aa-1-meta.nlm... Parsing 2024AA/META/MRSTY.RRF.gz as MRSTY with encoding UTF-8 Parsing 2024AA/META/MRRANK.RRF.gz as MRRANK with encoding UTF-8 Parsing 2024AA/META/MRCONSO.RRF.aa.gz as MRCONSO with encoding UTF-8 Parsing 2024AA/META/MRCONSO.RRF.ab.gz as MRCONSO with encoding UTF-8 Parsing 2024AA/META/MRDEF.RRF.gz as MRDEF with encoding UTF-8 Full UMLS release - importing UMLS from inner Zip file 2024AA-full/2024aa-2-meta.nlm... Parsing 2024AA/META/MRREL.RRF.aa.gz as MRREL with encoding UTF-8 Parsing 2024AA/META/MRREL.RRF.ab.gz as MRREL with encoding UTF-8 Parsing 2024AA/META/MRREL.RRF.ac.gz as MRREL with encoding UTF-8 Traceback (most recent call last): File "/home/mukun/projects/umls/test/symp_cui.py", line 45, in <module> import_umls("/mnt/c/Users/mukun/Downloads/umls-2024AA-full.zip", terminologies=["CUI", "SNOMEDCT_US", "ICD10"]) File "/home/mukun/.local/lib/python3.10/site-packages/owlready2/pymedtermino2/umls.py", line 710, in import_umls parse_umls_zip(umls_inner_zip) File "/home/mukun/.local/lib/python3.10/site-packages/owlready2/pymedtermino2/umls.py", line 684, in parse_umls_zip remnants[table_name] = parser(PYM, terminologies, langs, importer, File "/home/mukun/.local/lib/python3.10/site-packages/owlready2/pymedtermino2/umls.py", line 178, in parse_mrrel remnant, previous = remnant_previous TypeError: cannot unpack non-iterable NoneType object |
|
Administrator
|
Hello,
I've just tested, and I successfully imported UMLS 2024AA with Python version 3.12.6 and Owlready version 2-0.47. Which version of Owlready are you using? Your console log is different from mine so I suspect that it is not the latest version. Jiba |
«
Return to Owlready
|
1 view|%1 views
| Free forum by Nabble | Edit this page |